The Electron Collection#
Warning
Not all examples on this page have been updated to r22 and ServiceX 3. Examples that have not been updated will be marked using a similar warning. These examples will be updated soon.
The electron collection is a lot like the jet collection other than there are working points (loose, medium, tight) that are defined by the Egamma working group.
Accessing the collection is similar, at first blush, to the jet collection.
from config import get_data
from config import sx_f
from func_adl_servicex_xaodr22 import FuncADLQueryPHYSLITE
import matplotlib.pyplot as plt
import awkward as ak
The default electron we fetch is the so-called MediumLHElectron electron with NonIso isolation.
query = FuncADLQueryPHYSLITE()
electrons_per_event = (query
.Select(lambda e: e.Electrons())
.Select(lambda electrons: {
'pt': electrons.Select(lambda j: j.pt() / 1000),
'eta': electrons.Select(lambda j: j.eta()),
'phi': electrons.Select(lambda j: j.phi()),
})
)
electron_data = get_data(electrons_per_event, sx_f)
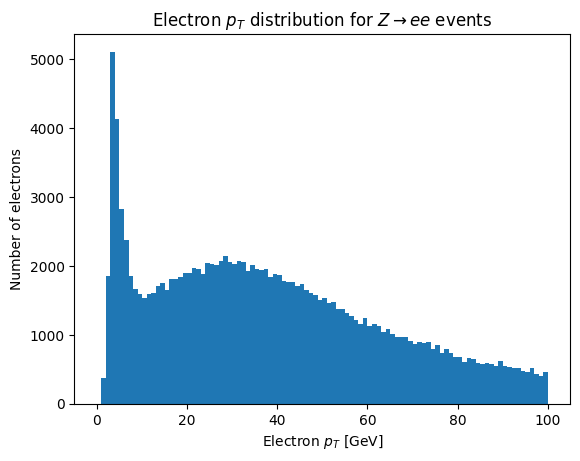
plt.hist(ak.flatten(electron_data.pt), bins=100, range=(0, 100))
plt.xlabel('Electron $p_T$ [GeV]')
plt.ylabel('Number of electrons')
_ = plt.title('Electron $p_T$ distribution for $Z\\rightarrow ee$ events')
Electron Types#
Warning
This example has not been updated to r22 and ServiceX 3 yet.
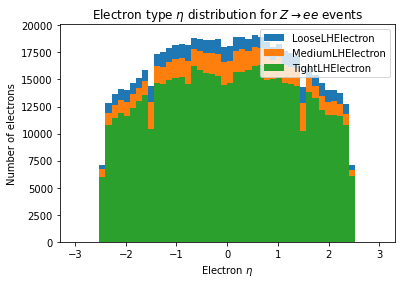
Electrons come in several different flavors. We can look at the different \(\eta\) distributions for the electrons. Isolation is another axis we can alter, not shown here, by changing the electron_isolation argument to Electrons.
In the calibration chapter we pointed out we can’t run multiple queries with different object setups due to limitations in the xAOD AnalysisBase when looking at calibrated objects. So we’ll have to do this in three queries instead. Also, we’ll have them run in parallel to improve efficiency a bit.
ele_working_points = ['LooseLHElectron', 'MediumLHElectron','TightLHElectron']
electrons_async = [
(calib_tools.query_update(ds, electron_working_point=wp)
.SelectMany(lambda e: [ele.eta() for ele in e.Electrons()])
.AsAwkwardArray('eta')
.value_async())
for wp in ele_working_points
]
electrons_wp = await asyncio.gather(*electrons_async)
---------------------------------------------------------------------------
NameError Traceback (most recent call last)
Cell In[4], line 2
1 ele_working_points = ['LooseLHElectron', 'MediumLHElectron','TightLHElectron']
----> 2 electrons_async = [
3 (calib_tools.query_update(ds, electron_working_point=wp)
4 .SelectMany(lambda e: [ele.eta() for ele in e.Electrons()])
5 .AsAwkwardArray('eta')
6 .value_async())
7 for wp in ele_working_points
8 ]
9 electrons_wp = await asyncio.gather(*electrons_async)
Cell In[4], line 3, in <listcomp>(.0)
1 ele_working_points = ['LooseLHElectron', 'MediumLHElectron','TightLHElectron']
2 electrons_async = [
----> 3 (calib_tools.query_update(ds, electron_working_point=wp)
4 .SelectMany(lambda e: [ele.eta() for ele in e.Electrons()])
5 .AsAwkwardArray('eta')
6 .value_async())
7 for wp in ele_working_points
8 ]
9 electrons_wp = await asyncio.gather(*electrons_async)
NameError: name 'calib_tools' is not defined
for idx, wp in enumerate(ele_working_points):
plt.hist(electrons_wp[idx].eta, bins=50, range=(-3, 3), label=wp)
plt.xlabel('Electron $\\eta$')
plt.ylabel('Number of electrons')
plt.title('Electron type $\\eta$ distribution for $Z\\rightarrow ee$ events')
_ = plt.legend()
Calibration#
Warning
This example has not been updated to r22 and ServiceX 3 yet.
By default the muons we pulled are MediumLHElectron quality and calibrated. We can grab the uncalibrated electrons easily enough:
To grab the raw jets (without calibration) we just set the calibrated parameter to False (there is very little reason one will do this normally):
raw_electrons = ( ds
.Select(lambda e: e.Electrons(uncalibrated_collection="Electrons"))
.Select(lambda eles: {
'pt': [e.pt() / 1000.0 for e in eles],
'eta': [e.eta() for e in eles],
'phi': [e.phi() for e in eles],
})
.AsAwkwardArray()
.value())
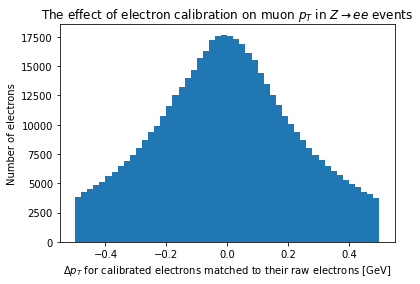
The number of raw jets and the number of calibrated jets are quite different from the number of raw jets, so we’ll need to match them in \(\eta\) and \(\phi\):
raw_electrons_matched = match_objects(electrons, raw_electrons)
And we can compared the matched electrons:
plt.hist(ak.flatten(electrons.pt-raw_electrons_matched.pt), bins=50, range=(-0.5, 0.5))
plt.xlabel(r'$\Delta p_T$ for calibrated electrons matched to their raw electrons [GeV]')
plt.ylabel('Number of electrons')
_ = plt.title('The effect of electron calibration on muon $p_T$ in $Z\\rightarrow ee$ events')
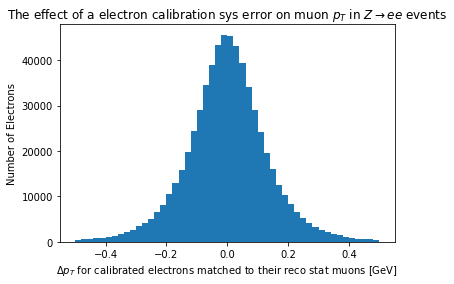
If we instead want a particular systematic error, we need only name that error to get it back. Knowing what the names of the systematic errors, however, is not something that can be programmatically determined ahead of time. See the further information section at the end of this chapter to links to the ATLAS electron calibration info twiki. Though I’ve noticed it is not necessarily up to date with names.
sys_electrons = (calib_tools.query_sys_error(ds, 'EG_RESOLUTION_ALL__1up')
.Select(lambda e: e.Electrons())
.Select(lambda eles: {
'pt': [e.pt() / 1000.0 for e in eles],
'eta': [e.eta() for e in eles],
'phi': [e.phi() for e in eles],
})
.AsAwkwardArray()
.value())
sys_electrons_match = match_objects(electrons, sys_electrons)
plt.hist(ak.flatten(electrons.pt-sys_electrons_match.pt), bins=50, range=(-0.5, 0.5))
plt.xlabel('$\Delta p_T$ for calibrated electrons matched to their reco stat muons [GeV]')
plt.ylabel('Number of Electrons')
_ = plt.title('The effect of a electron calibration sys error on muon $p_T$ in $Z\\rightarrow ee$ events')
The Data Model#
The Electron_v1 class at the time this documentation was built:
from func_adl_servicex_xaodr22.xAOD.electron_v1 import Electron_v1
help(Electron_v1)
Help on class Electron_v1 in module func_adl_servicex_xaodr22.xAOD.electron_v1:
class Electron_v1(builtins.object)
| A class
|
| Methods defined here:
|
| OQ(self) -> 'int'
| A method
|
| ambiguousObject(self) -> 'func_adl_servicex_xaodr22.xAOD.egamma_v1.Egamma_v1'
| A method
|
| caloCluster(self, index: 'int') -> 'func_adl_servicex_xaodr22.xAOD.calocluster_v1.CaloCluster_v1'
| A method
|
| caloClusterLink(self, index: 'int') -> 'func_adl_servicex_xaodr22.elementlink_datavector_xaod_calocluster_v1__.ElementLink_DataVector_xAOD_CaloCluster_v1__'
| A method
|
| caloClusterLinks(self) -> 'func_adl_servicex_xaodr22.vector_elementlink_datavector_xaod_calocluster_v1___.vector_ElementLink_DataVector_xAOD_CaloCluster_v1___'
| A method
|
| charge(self) -> 'float'
| A method
|
| clearDecorations(self) -> 'bool'
| A method
|
| e(self) -> 'float'
| A method
|
| eta(self) -> 'float'
| A method
|
| hasNonConstStore(self) -> 'bool'
| A method
|
| hasStore(self) -> 'bool'
| A method
|
| index(self) -> 'int'
| A method
|
| isGoodOQ(self, mask: 'int') -> 'bool'
| A method
|
| likelihoodValue(self, value: 'float', LHValue: 'func_adl_servicex_xaodr22.str.str') -> 'bool'
| A method
|
| m(self) -> 'float'
| A method
|
| nCaloClusters(self) -> 'int'
| A method
|
| nTrackParticles(self) -> 'int'
| A method
|
| p4(self) -> 'func_adl_servicex_xaodr22.tlorentzvector.TLorentzVector'
| A method
|
| passSelection(self, value: 'bool', menu: 'func_adl_servicex_xaodr22.str.str') -> 'bool'
| A method
|
| phi(self) -> 'float'
| A method
|
| pt(self) -> 'float'
| A method
|
| rapidity(self) -> 'float'
| A method
|
| selectionisEM(self, value: 'int', isEM: 'func_adl_servicex_xaodr22.str.str') -> 'bool'
| A method
|
| trackIndices(self) -> 'bool'
| A method
|
| trackParticle(self, index: 'int') -> 'func_adl_servicex_xaodr22.xAOD.trackparticle_v1.TrackParticle_v1'
| A method
|
| trackParticleLink(self, index: 'int') -> 'func_adl_servicex_xaodr22.elementlink_datavector_xaod_trackparticle_v1__.ElementLink_DataVector_xAOD_TrackParticle_v1__'
| A method
|
| trackParticleLinks(self) -> 'func_adl_servicex_xaodr22.vector_elementlink_datavector_xaod_trackparticle_v1___.vector_ElementLink_DataVector_xAOD_TrackParticle_v1___'
| A method
|
| usingPrivateStore(self) -> 'bool'
| A method
|
| usingStandaloneStore(self) -> 'bool'
| A method
|
| ----------------------------------------------------------------------
| Readonly properties defined here:
|
| auxdataConst
| A method
|
| isAvailable
| A method
|
| ----------------------------------------------------------------------
| Data descriptors defined here:
|
| __dict__
| dictionary for instance variables (if defined)
|
| __weakref__
| list of weak references to the object (if defined)
Further Information#
The
xAOD::Electron_v1C++ header file with all the inline documentation.The Electron Recommendation Pages for R21 on the ATLAS TWiki
The Electron Working Points Pages for R21 on the ATLAS Twiki
